🔧 Cloud-Optimized Data Access#
Recall from the Cloud Data Access Notebook that cloud object storage is accessed over the network. Local file storage access will always be faster but there are limitations. This is why the design of file formats in the cloud requires more consideration than local file storage.
🤔 What is one limitation of local file storage?
See the table Cloud vs Local Storage in the Cloud Data Access Notebook.
Why you should care#
Reading ICESat-2 files, which are often large and not cloud-optimized, can be slow! It is nice to know why and possibly advocate for things to be better!
What are we optimizing for and why?#
The “optimize” in cloud-optimized is to minimize data latency and maximize throughput (see glossary below) by:
Making as few requests as possible;
Making even less for metadata, preferably only one; and
Using a file layout that simplifies accessing data for parallel reads.
A future without file formats
I like to imagine a day when we won’t have to think about file formats. The geospatial software community is working on ways to make all collections appear as logical datasets, so you can query them without having to think about files.
Anatomy of a structured data file#
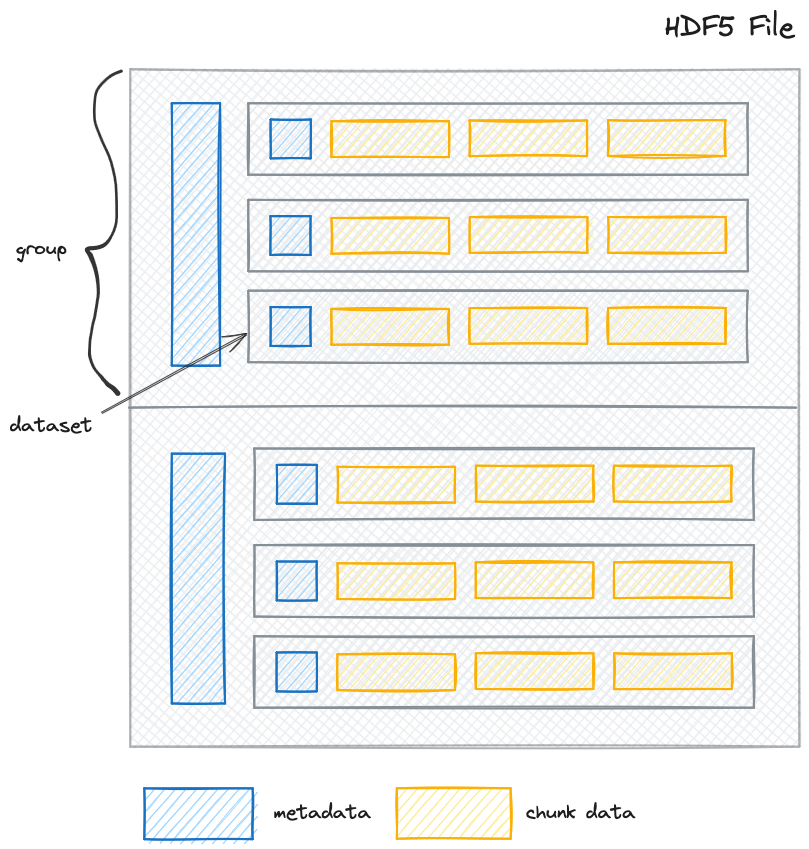
A structured data file is composed of two parts: metadata and the raw data. Metadata is information about the data, such as the data shape, data type, the data variables, the data's coordinate system, and how the data is stored, such as chunk shape and compression. It is also crucial to **lazy loading** (see glossary belo). Data is the actual data that you want to analyze. Many geospatial file formats, such as GeoTIFF, are composed of metadata and data.
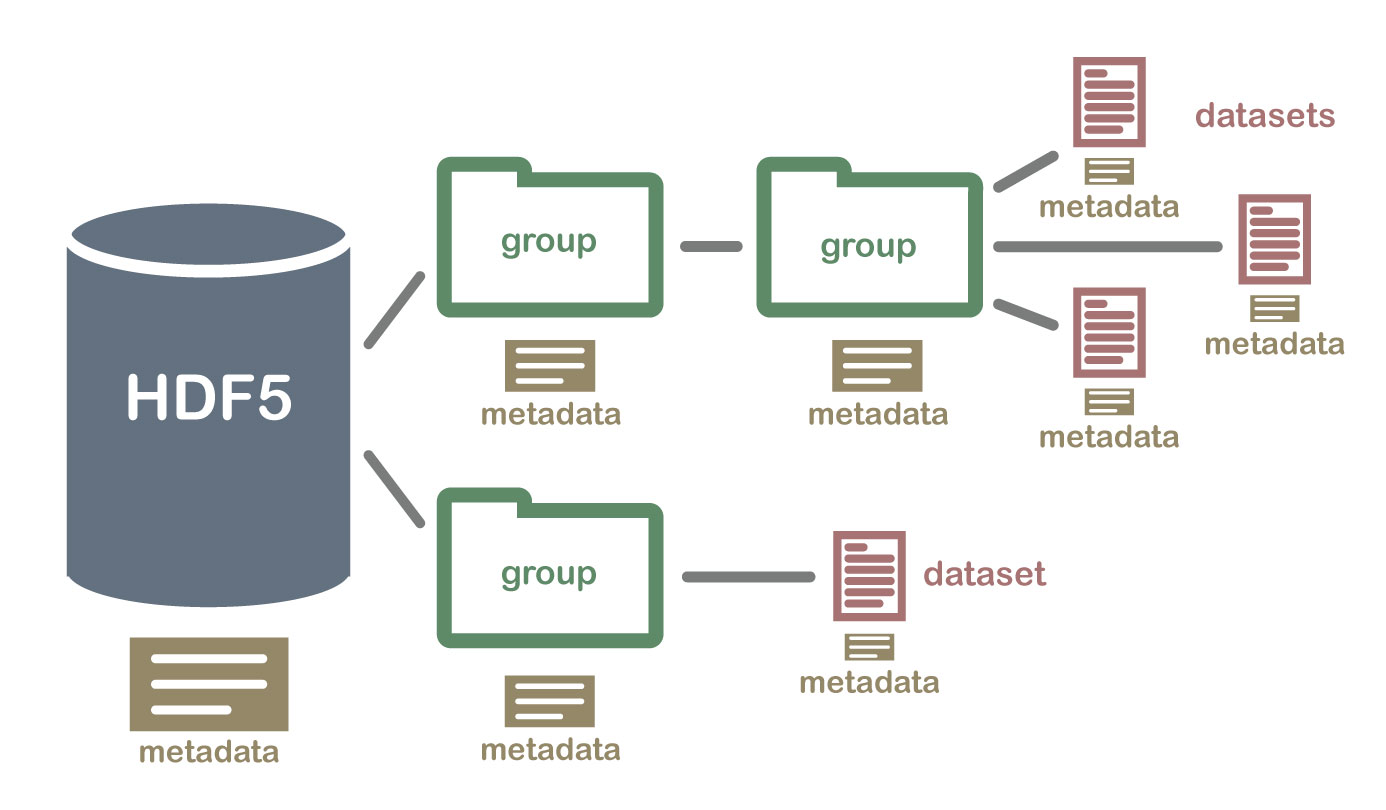
Image source: https://www.neonscience.org/resources/learning-hub/tutorials/about-hdf5
We can optimize this structure for reading from cloud storage.
How should we structure files for the cloud?#
A “moving away from home” analogy#
Imagine when you lived at home with your parents. Everything was right there when you needed it (like local file storage). Let’s say you’re about to move away to college (the cloud), but you have decided to backpack there and so you can’t bring any of your belongings with you. You put everything in your parent’s (infinitely large) garage (cloud object storage). Given you would need to have things shipped to you, would it be better to leave everything unpacked? To put everything all in one box? A few different boxes? And what would be the most efficient way for your parents to know where things were when you asked for them?

image generated with ChatGPT 4
You can actually make any common geospatial data formats (HDF5/NetCDF, GeoTIFF, LAS (LIDAR Aerial Survey)) “cloud-optimized” by:
Separate metadata from data and store metadata contiguously so it can be read with one request.
Store data in chunks, so the whole file doesn’t have to be read to access a portion of the data, and it can be compressed.
Make sure the chunks of data are not too small, so more data is fetched with each request.
Make sure the chunks are not too large, which means more data has to be transferred and decompression takes longer.
Compress these chunks so there is less data to transfer over the network.
Glossary#
latency#
The time between when data is sent to when it is received. Read more.
throughput#
The amount of data that can be transferred over a given time. Read more.
lazy loading#
🛋️ 🥔 Lazy loading is deferring loading any data until required. Here’s how it works: Metadata stores a mapping of chunk indices to byte ranges in files. Libraries, such as xarray, read only the metadata when opening a dataset. Libraries defer requesting any data until values are required for computation. When a computation of the data is finally called, libraries use HTTP range requests to request only the byte ranges associated with the data chunks required. See the s3fs cat_ranges function and xarray’s documentation on lazy indexing.
Opening Arguments
A few arguments used to open the dataset also make a huge difference, namely with how libraries, such as s3fs and h5py, cache chunks.
For s3fs, use cache_type and block_size.
For h5py, use rdcc_nbytes and page_buf_size.
See also
Cloud-Optimized HDF5 Files – Aleksandar Jelenak, The HDF Group
HDF at the speed of Zarr - Luis Lopez, Pangeo Showcase is a presentation all about Cloud-Optimizing ICESat-2 Products
A notebook demonstrating how to repack ATL03 product to cloud-optimized (for a subset of datasets): rechunking_atl03.ipynb
Takeaways
Understanding file formats may help in diagnosing issues when things are slow.
You can make files cloud-optimized by separating metadata and storing it contiguously so it can all be read in one request.
You can use arguments to libraries like s3fs and h5py to support caching.

