Making Data Interoperable and Reusable#
We want to make sure that our data can be used by other researchers as easily as possible.
Ask yourself the question, if I dissappear for a month, can someone else open my data files, know what the data is, and start using the data?
The FAIR principles layout systematic requirements to make data Findable, Accessible, Interoperable and Reusable. In this section we will focus on the Interoperable and Reusable parts of FAIR.
What is Interoperable data#
Interoperability means that data can be used by applications and in workflows for analysis, and can be integrated with other data.
I tend to think of interoperability as the number of steps (or headaches) required to read, transform and plot data. A useful guide is to ask the following questions about your dataset.
Is the data in a standard file format?#
Fortunately, the Earth science community has, mostly, converged on a standard set of well defined file formats. Common traditional and cloud-optimized geospatial file formats for vector, point-cloud and N-dimensional data.Source: Cloud-optimized geospatial formats guide shows traditional and cloud-optimized formats for some common types of geospatial data. See the cloud optimized data tutorial for more information of cloud-optimized file formats. This tutorial will focus on traditional formats but a lot of what is covered also applies to cloud-optimized formats.
Note
Geospatial data is any data that can be referenced to a location on Earth; either at the surface or above or below the surface.
The choice of which file format to store your data in can be guided the type of data you need to store.
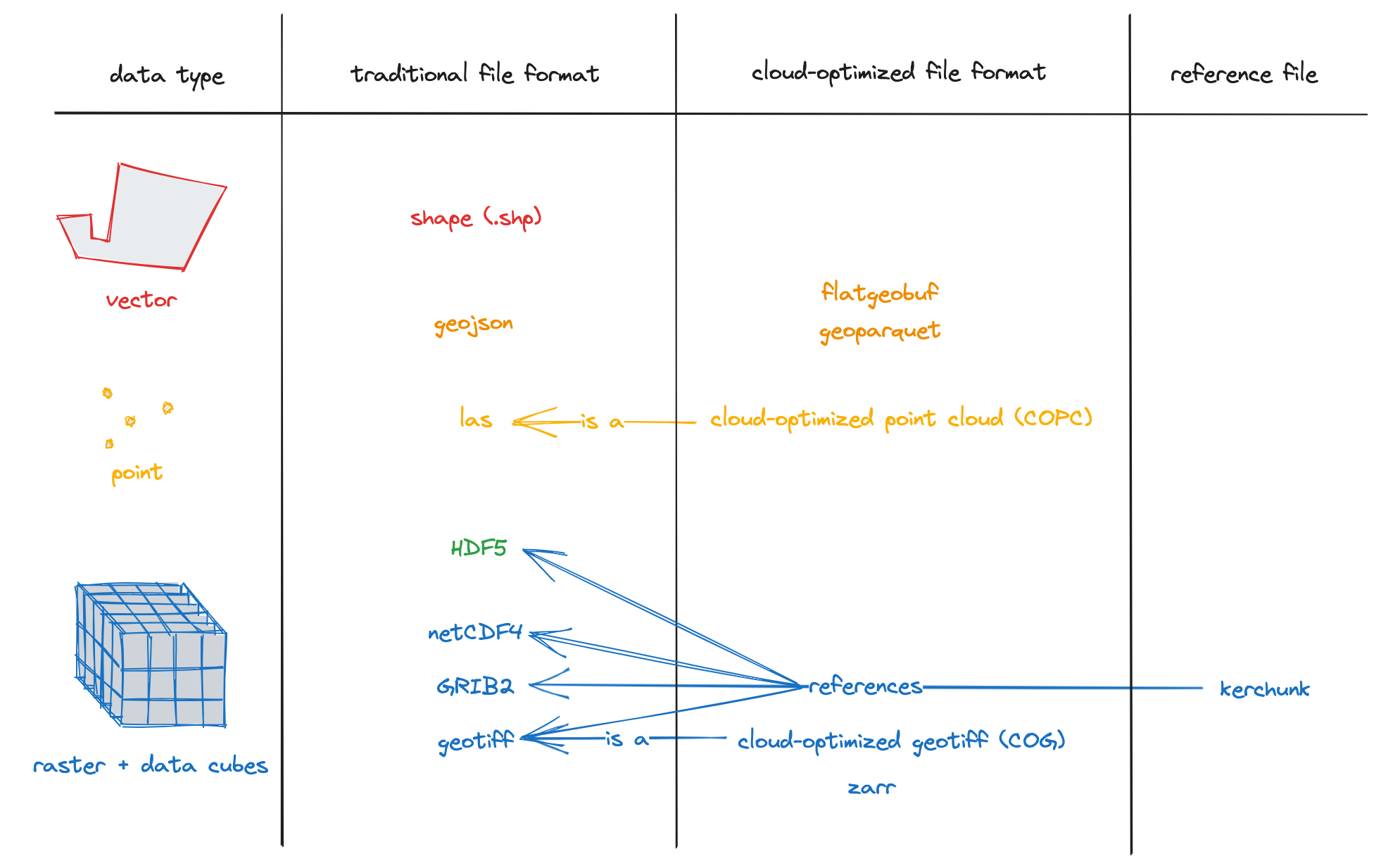
Fig. 2 Common traditional and cloud-optimized geospatial file formats for vector, point-cloud and N-dimensional data.
Source: Cloud-optimized geospatial formats guide#
Vector data is data represented as points, lines and polygons. Each vetor entity can have attributes associated with it. For example, a network of weather stations would be represented as a set of points. A ship track or buoy trajectory would be represented as a line. A set of glacier outlines are a set of polygons. Vector data are commonly stored as Shapefiles or GeoJSON.
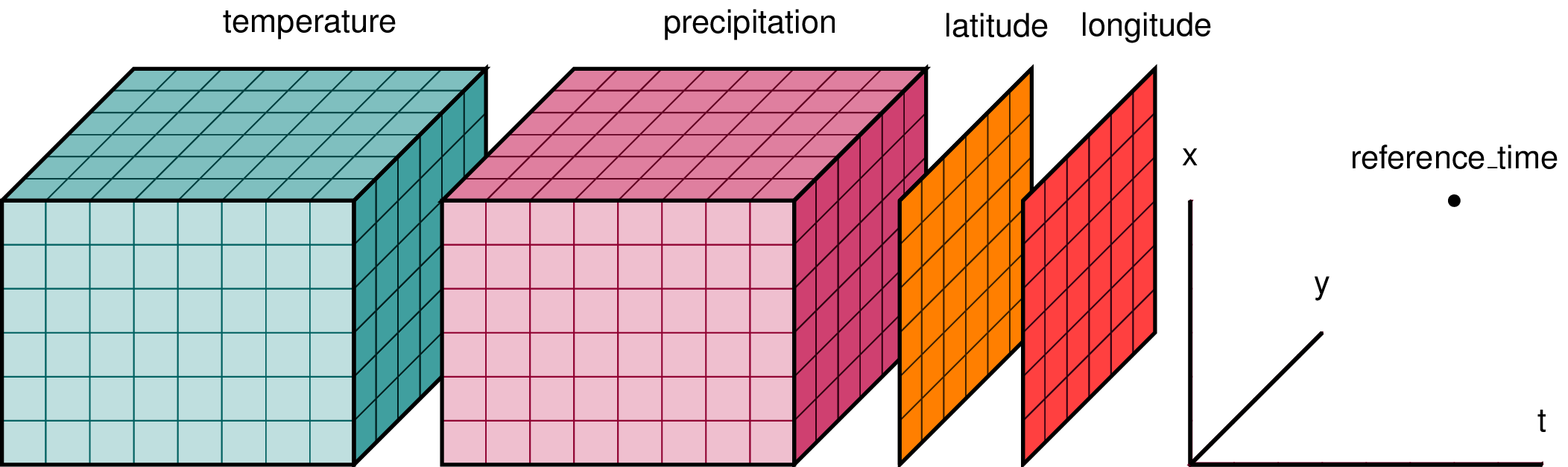
Fig. 3 A cartoon of a NetCDF-style N-dimensional data structure containing (\(time\),\(x\),\(y\)) data-cubes of air temperature and precipitation, and latitude and longitude coordinate variables. The named dimensions in the file are shown as orthogonal axes to the right of the variables. Source: xarray-dev#
N-dimensional data is data with two or more dimensions. Two examples are a geolocated remote sensing image, and a data-cube (\(t\),\(x\),\(y\)) of temperature grids from a climate model (e.g. A cartoon of a NetCDF-style N-dimensional data structure containing (time,x,y) data-cubes of air temperature and precipitation, and latitude and longitude coordinate variables. The named dimensions in the file are shown as orthogonal axes to the right of the variables. Source: xarray-dev). NetCDF4 is commonly used to store N-dimensional datasets because it allows coordinates to be stored with the data variables so that data can be located in space and time, as well as allowing attributes for each data variable to be stored. Global attributes can also be stored with each file. This makes the format self-describing, which adds to interoperability and reusability.
The standard format for geolocated image data is GeoTIFF (Geolocated Tagged Image File Format). GeoTIFF stores information that allows each image pixel to be located on Earth. Multiple bands can be stored. However, GeoTIFF files cannot store the rich metadata that NetCDF files can store.
Point cloud data are three-dimensional data (\(x\),\(y\),\(z\)) with attributes usually produced by Lidar or photogrammetry. This could be a mapping of a snow surface from Terrestrial Scanning Lidar. Point cloud data shares characteristics with Vector data but generall has a much higher density of points.
A fourth type of data is Tabular data, which is data structured as rows and columns. Weather station records are tabular data. The data below contains air temperature and dewpoint temperature data recorded at 5-minute intervals for the Taos, NM, Soil and Water Conservation District site.
datetime, air_temperature, dewpoint
2024-08-12 16:05:00, 87.2, 55.5
2024-08-12 16:10:00, 86.7, 55.6
2024-08-12 16:15:00, 86.4, 55.5
2024-08-12 16:20:00, 87.3, 55.2
2024-08-12 16:25:00, 86.8, 54.8
2024-08-12 16:30:00, 87.6, 55.1
Often this data can be stored as a Comma Seperated File (csv). For small volumes of data csv is fine. Data are both machine and human readable, which is great for checking data by hand. NetCDF or even HDF5 could be used and would allow attributes to be included. Cloud optimized formats such as Parquet could also be used.
Hierachical Data Format version 5 (HDF5) is widely used by NASA, and other to store complex data structures. ICESat-2 data is one example. The Geolocated Photon Height product (ATL03) contains over 1000 variables. These variables are organized into groups with different dimension sizes. Each variable has a set of attributes associated with it.
Can the data be read by common tools?#
Putting data into the common standard file formats used in your discipline will make it more likely that users of your data can read the data with commonly used tools. I will focus on Python tools here but R, Julia, and Rust languages, and Matlab and IDL all have ways to read HDF5, NetCDF, GeoTIFF, Shapefiles, GeoJSON, and csv.
The common Python tools used for working with geospatial and tabular data are listed in the table below.
Datatype |
Formats |
Tools |
|---|---|---|
Vector |
Shapefile |
|
GeoJSON |
|
|
N-dimensional data |
NetCDF |
|
GeoTIFF |
|
|
HDF5 |
|
|
Point cloud |
las |
?? |
Tabular Data |
csv |
|
Most, if not all, of these tools have write methods to create files in the described in standard file formats
Does the data follow domain standards and conventions#
Storing data in standardized data structures and file formats that data can be read and manipulated using a standard set of tools. By following domain specific conventions and “best practices”, software packages and machines, and users, can understand (interpret) data.
Climate Forecast (CF) Conventions are relevant for most N-dimensional data. The purpose of CF conventions is to make NetCDF files self describing.
{attribution=”NetCDF Climate and Forecast (CF) Metadata Conventions, Version 1.11”}
The purpose of the CF conventions is to require conforming datasets to contain sufficient metadata that they are self-describing in the sense that each variable in the file has an associated description of what it represents, including physical units if appropriate, and that each value can be located in space (relative to earth-based coordinates) and time.
An important benefit of a convention is that it enables software tools to display data and perform operations on specified subsets of the data with minimal user intervention.
The dump of a subsetted ERA5 Reanalaysis file shows the structure of a CF-compliant file
netcdf era5.single_levels.monthly.1959 {
dimensions:
longitude = 1440 ;
latitude = 721 ;
time = 12 ;
variables:
float longitude(longitude) ;
longitude:units = "degrees_east" ;
longitude:long_name = "longitude" ;
float latitude(latitude) ;
latitude:units = "degrees_north" ;
latitude:long_name = "latitude" ;
int time(time) ;
time:units = "hours since 1900-01-01 00:00:00.0" ;
time:long_name = "time" ;
time:calendar = "gregorian" ;
short t2m(time, latitude, longitude) ;
t2m:scale_factor = 0.00166003112108415 ;
t2m:add_offset = 258.629197755436 ;
t2m:_FillValue = -32767s ;
t2m:missing_value = -32767s ;
t2m:units = "K" ;
t2m:long_name = "2 metre temperature" ;
short tcwv(time, latitude, longitude) ;
tcwv:scale_factor = 0.00104011870543087 ;
tcwv:add_offset = 34.1879291446153 ;
tcwv:_FillValue = -32767s ;
tcwv:missing_value = -32767s ;
tcwv:units = "kg m**-2" ;
tcwv:long_name = "Total column vertically-integrated water vapour" ;
tcwv:standard_name = "lwe_thickness_of_atmosphere_mass_content_of_water_vapor" ;
// global attributes:
:Conventions = "CF-1.6" ;
:history = "2022-06-17 18:39:10 GMT by grib_to_netcdf-2.24.3: /opt/ecmwf/mars-client/bin/grib_to_netcdf -S param -o /cache/data7/adaptor.mars.internal-1655491150.0401924-522-6-56f5a114-0c72-40e2-bb43-9866e22ab084.nc /cache/tmp/56f5a114-0c72-40e2-bb43-9866e22ab084-adaptor.mars.internal-1655491144.161893-522-11-tmp.grib" ;
}
NetCDF has the concept of two types of variables: data variables and coordinate variables. In this example, longitude, latitude and time are all coordinate variables. The coordinate variables are 1-dimensional and have the same names as the three dimensions of the file. The data variables are t2m and tcwv. These are linked to the dimensions and coordinate variables.
Note
Having 1-dimensional coordinate variables is really helpful because you can subset data by coordinate values easily. Add an example for this
Both coordinate variables and data variables have attributes that describe what each variable is and what the units are. These are critical for both humans and software to understand what the data are. There are also global attributes that are used to describe the conventions used, here CF Conventions, version 1.6, and processing history.
CF Conventions require spatial coordinate variables (longitude and latitude in this case) to have units and a long_name attribute. degrees_north and degrees_east are the recommended units for for latitude and longitude. If data are in projected coordinates (e.g. x and y), these woud have units m for meters.
The time variable also has units, a long_name and a calendar attribute. The units have the format interval since reference_time where interval can be seconds, minutes, hours and days.
years and months are allowed by not recommended because the length or a year and a month can vary.
days since 1971-04-21 06:30:00.0
A calendar is important because it defines how time is incremented. Some climate models use 360-day or 365-day calendars with no leap year. Paleo-data may used a mixed Julian/Gregorian or proleptic Gregorian calendar, in which dates before the switch from Julian to Gregorian calendars follow the Gregorian leap-year rules.
Try to avoid a year zero and remember that months and days start at one not zero!
Does the file contain useful coordinates (time, x, y, z)?#
Geospatial data needs to be located on Earth and to a date. So data needs coordinates. Spatial coordinates may be latitude, longitude and height (or depth), or x, y, and z usually in meters. But heights may be pressure levels, sigma levels, density, potential temperature.
Does the file have a coordinate reference system?#
Coordinate values are associated with a set of axes or coordinate system. A Coordinate Reference System (CRS) is required to relate the coordinate system to Earth. Even if your data is in latitude and longitude it is better to be explicit and give the CRS. It is not always wise to assume that latitudes and longitudes are in WGS84 (EPSG:4326).
Do the variables have standard names, units, etc?#
Units are just as important now as they were for your physics homework. Standard names define your data in a standard way so that users know exactly what the variable is.
What is Reusable data#
Reusability means that the data have sufficient information about what it is, how it was collected and processed so that future users can understand the data and use it correctly.
I think of this as having most of my questions answered about the data.
Is it appropriate to use this data for what I want to do?
Is the data structured for analysis?#
In most cases, vector data are structured so that they can be used in analysis workflows. However, n-dimensional data and tabular data are often messy and need to be tidied before you can do analysis. Below is a list of some ways to make your data tidy.
Note
Tidy data is a term from the R community, popularized by Hadley Wickham, for tabular data that is structured for easy analysis. Here, I also apply it to n-dimensional data.
Warning
While many of these “best practices” are documented by other data managers and users elsewhere, some are my own opinionated “best practice”. I make no apologies for this. Where possible, I give what I hope is a good reason for the practice. Where I give no reason, it is probably because at some point I got annoyed with having to spend hours cleaning data and the “best practice” could have saved me time.
“Tidy” n-dimensional data#
Data variables should contain only data values
Try to avoid using groups if at all possible -
xarraycannot read multiple groups but seeDatatree
Tidy tabular data#
Above all remember that your data needs to be read by a machine, and machines are dumb! No matter what ChatGPT or Elon Musk tells you. You may be able to read a nicely formatted page in Excel but a computer will have a hard time, if not find it impossible.
rows are records - e.g. measurements for a single location, timestep or class - show example
columns are a single variable
Prefer iso-standard datetimes rather than breaking date components into separate columns. Users (including a future you) will only have to reconstruct the datetimes from these columns. And tools like
pandascan select by year, month and day automatically.Do not include marginal sums and statistics in the data
Avoid using spaces in column names

