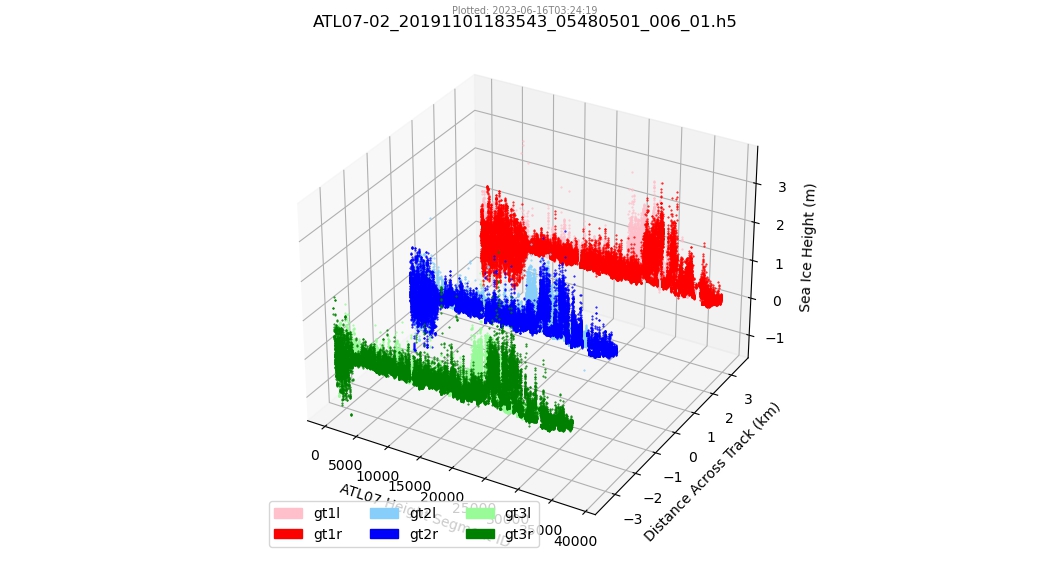
Machine Learning with ICESat-2 data#
A machine learning pipeline for point cloud classification.
Reimplementation of Koo et al., 2023, based on code available at YoungHyunKoo/IS2_ML.
Learning Objectives
By the end of this tutorial, you should be able to:
Convert ICESat-2 point cloud data into an analysis/ML-ready format
Recognize the different levels of complexity of ML approaches and the benefits/challenges of each
Learn the potential of using ML for ICESat-2 point cloud classification
Part 0: Setup#
Let’s start by importing all the libraries needed for data access, processing and visualization later. If you’re running this on CryoCloud’s default image without Pytorch installed, uncomment and run the first cell before continuing.
# !mamba install -y pytorch
import earthaccess
import geopandas as gpd
import h5py
import numpy as np
import pandas as pd
import pygmt
import pystac_client
import rioxarray # noqa: F401
import shapely.geometry
import stackstac
import torch
import tqdm
Part 1: Convert ICESat-2 data into ML-ready format#
Steps:
Get ATL07 data using earthaccess
Find coincident Sentinel-2 imagery by searching over a STAC API
Filter to only strong beams, and 6 key data variables + ancillary variables
Search for ATL07 data over study area#
In this sub-section, we’ll set up a spatiotemporal query to look for ICESat-2 ATL07 sea ice data over the Ross Sea region around late October 2019.
Ref: https://earthaccess.readthedocs.io/en/latest/quick-start/#get-data-in-3-steps
# Authenticate using NASA EarthData login
auth = earthaccess.login()
# s3 = earthaccess.get_s3fs_session(daac="NSIDC") # Start an AWS S3 session
# Set up spatiotemporal query for ATL07 sea ice product
granules = earthaccess.search_data(
short_name="ATL07",
cloud_hosted=True,
bounding_box=(-180, -78, -140, -70), # xmin, ymin, xmax, ymax
temporal=("2019-10-31", "2019-11-01"),
version="006",
)
granules[-1] # visualize last data granule
Find coincident Sentinel-2 imagery#
Let’s also find some optical satellite images that were captured at around the same
time and location as the ICESat-2 ATL07 tracks. We will be using
pystac_client.Client.search
and doing the search with two steps:
(Fast) search using date to find potential Sentinel-2/ICESat-2 pairs
(Slow) search using spatial intersection to ensure Sentinel-2 image overlaps with ICESat-2 track.
# Connect to STAC API that hosts Sentinel-2 imagery on AWS us-west-2
catalog = pystac_client.Client.open(url="https://earth-search.aws.element84.com/v1")
# Loop over each ATL07 data granule, and find Sentinel-2 images from same time range
for granule in tqdm.tqdm(iterable=granules):
# Get ATL07 time range from Unified Metadata Model (UMM)
date_range = granule["umm"]["TemporalExtent"]["RangeDateTime"]
start_time = date_range["BeginningDateTime"] # e.g. '2019-02-24T19:51:47.580Z'
end_time = date_range["EndingDateTime"] # e.g. '2019-02-24T19:52:08.298Z'
# 1st check (temporal match)
search1 = catalog.search(
collections="sentinel-2-l2a", # Bottom-of-Atmosphere product
bbox=[-180, -78, -140, -70], # xmin, ymin, xmax, ymax
datetime=f"{start_time}/{end_time}",
query={"eo:cloud_cover": {"lt": 30}}, # max cloud cover
max_items=10,
)
_item_collection = search1.item_collection()
if (_item_len := len(_item_collection)) >= 1:
# print(f"Potential: {_item_len} Sentinel-2 x ATL07 matches!")
# 2nd check (spatial match) using centre-line track intersection
file_obj = earthaccess.open(granules=[granule])[0]
atl_file = h5py.File(name=file_obj, mode="r")
linetrack = shapely.geometry.LineString(
coordinates=zip(
atl_file["gt2r/sea_ice_segments/longitude"][:10000],
atl_file["gt2r/sea_ice_segments/latitude"][:10000],
)
).simplify(tolerance=10)
search2 = catalog.search(
collections="sentinel-2-l2a",
intersects=linetrack,
datetime=f"{start_time}/{end_time}",
max_items=10,
)
item_collection = search2.item_collection()
if (item_len := len(item_collection)) >= 1:
print(
f"Found: {item_len} Sentinel-2 items coincident with granule:\n{granule}"
)
break # uncomment this line if you want to find more matches
25%|██▌ | 2/8 [00:00<00:00, 12.12it/s]
38%|███▊ | 3/8 [00:03<00:05, 1.12s/it]
Found: 4 Sentinel-2 items coincident with granule:
Collection: {'EntryTitle': 'ATLAS/ICESat-2 L3A Sea Ice Height V006'}
Spatial coverage: {'HorizontalSpatialDomain': {'Orbit': {'AscendingCrossing': 34.39032297860235, 'StartLatitude': -27.0, 'StartDirection': 'D', 'EndLatitude': -27.0, 'EndDirection': 'A'}}}
Temporal coverage: {'RangeDateTime': {'BeginningDateTime': '2019-10-31T20:05:15.872Z', 'EndingDateTime': '2019-10-31T20:20:05.522Z'}}
Size(MB): 57.1460075378418
Data: ['https://data.nsidc.earthdatacloud.nasa.gov/nsidc-cumulus-prod-protected/ATLAS/ATL07/006/2019/10/31/ATL07-02_20191031190122_05330501_006_01.h5']
We should have found a match! In case you missed it, these are the two variables pointing to the data we’ll use later:
granule- ICESat-2 ATL07 sea ice point cloud dataitem_collection- Sentinel-2 optical satellite images
Filter to strong beams and required data variables#
Here, we’ll open one ATL07 sea ice data file, and do some pre-processing.
%%time
file_obj = earthaccess.open(granules=[granule])[0]
atl_file = h5py.File(name=file_obj, mode="r")
atl_file.keys()
CPU times: user 51.8 ms, sys: 7.77 ms, total: 59.5 ms
Wall time: 148 ms
<KeysViewHDF5 ['METADATA', 'ancillary_data', 'gt1l', 'gt1r', 'gt2l', 'gt2r', 'gt3l', 'gt3r', 'orbit_info', 'quality_assessment']>
Strong beams can be chosen based on the sc_orient variable.
Ref: ICESAT-2HackWeek/strong-beams
# orientation - 0: backward, 1: forward, 2: transition
orient = atl_file["orbit_info"]["sc_orient"][:]
if orient == 0:
strong_beams = ["gt1l", "gt2l", "gt3l"]
elif orient == 1:
strong_beams = ["gt3r", "gt2r", "gt1r"]
strong_beams
['gt3r', 'gt2r', 'gt1r']
To keep things simple, we’ll only read one beam today, but feel free to get all three using a for-loop in your own project.
for beam in strong_beams:
print(beam)
gt3r
gt2r
gt1r
Key data variables to use (for model training):
photon_rate: photon ratehist_w: width of the photon height distributionbackground_r_norm: background photon rateheight_segment_height: relative surface heightheight_segment_n_pulse_seg: number of laser pulseshist_mean_median_h_diff=hist_mean_h-hist_median_h: difference between mean and median height
Other data variables:
x_atc- Along track distance from the equatorlayer_flag- Consolidated cloud flag { 0: ‘likely_clear’, 1: ‘likely_cloudy’ }height_segment_ssh_flag- Sea surface flag { 0: ‘sea ice’, 1: ‘sea surface’ }
Data dictionary at: https://nsidc.org/sites/default/files/documents/technical-reference/icesat2_atl07_data_dict_v006.pdf
gdf = gpd.GeoDataFrame(
data={
# Key data variables
"photon_rate": atl_file[f"{beam}/sea_ice_segments/stats/photon_rate"][:],
"hist_w": atl_file[f"{beam}/sea_ice_segments/stats/hist_w"][:],
"background_r_norm": atl_file[
f"{beam}/sea_ice_segments/stats/background_r_norm"
][:],
"height_segment_height": atl_file[
f"{beam}/sea_ice_segments/heights/height_segment_height"
][:],
"height_segment_n_pulse_seg": atl_file[
f"{beam}/sea_ice_segments/heights/height_segment_n_pulse_seg"
][:],
"hist_mean_h": atl_file[f"{beam}/sea_ice_segments/stats/hist_mean_h"][:],
"hist_median_h": atl_file[f"{beam}/sea_ice_segments/stats/hist_median_h"][:],
# Other data variables
"x_atc": atl_file[f"{beam}/sea_ice_segments/seg_dist_x"][:],
"layer_flag": atl_file[f"{beam}/sea_ice_segments/stats/layer_flag"][:],
"height_segment_ssh_flag": atl_file[
f"{beam}/sea_ice_segments/heights/height_segment_ssh_flag"
][:],
},
geometry=gpd.points_from_xy(
x=atl_file[f"{beam}/sea_ice_segments/longitude"][:],
y=atl_file[f"{beam}/sea_ice_segments/latitude"][:],
),
crs="OGC:CRS84",
)
# Pre-processing data
gdf = gdf[gdf.layer_flag == 0].reset_index(drop=True) # keep non-cloudy points only
gdf["hist_mean_median_h_diff"] = gdf.hist_mean_h - gdf.hist_median_h
print(f"Total number of rows: {len(gdf)}")
Total number of rows: 38246
gdf
| photon_rate | hist_w | background_r_norm | height_segment_height | height_segment_n_pulse_seg | hist_mean_h | hist_median_h | x_atc | layer_flag | height_segment_ssh_flag | geometry | hist_mean_median_h_diff | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.512820 | 0.128929 | 3470228.250 | 0.085903 | 38 | -53.941635 | -53.943508 | 2.744329e+07 | 0 | 0 | POINT (-166.17149 -66.1212) | 0.001873 |
| 1 | 4.181818 | 0.159623 | 3462216.000 | 0.117592 | 32 | -53.906929 | -53.901737 | 2.744330e+07 | 0 | 0 | POINT (-166.17152 -66.12129) | -0.005192 |
| 2 | 4.156250 | 0.189304 | 3460210.250 | 0.170189 | 31 | -53.872688 | -53.853218 | 2.744331e+07 | 0 | 0 | POINT (-166.17154 -66.12139) | -0.019470 |
| 3 | 4.551724 | 0.272234 | 3457500.000 | 0.236546 | 28 | -53.877270 | -53.831566 | 2.744332e+07 | 0 | 0 | POINT (-166.17156 -66.12148) | -0.045704 |
| 4 | 4.620690 | 0.224559 | 3411361.250 | 0.252434 | 28 | -53.828976 | -53.795189 | 2.744333e+07 | 0 | 0 | POINT (-166.17159 -66.12156) | -0.033787 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 38241 | 7.722222 | 0.146530 | 1436920.250 | 0.075413 | 17 | 18.942928 | 18.955540 | 3.303276e+07 | 0 | 0 | POINT (18.94094 -63.78423) | -0.012611 |
| 38242 | 7.944445 | 0.142120 | 1436920.250 | 0.048256 | 17 | 18.921795 | 18.934738 | 3.303276e+07 | 0 | 0 | POINT (18.94093 -63.78418) | -0.012943 |
| 38243 | 7.578948 | 0.140504 | 1436920.250 | 0.033785 | 18 | 18.893675 | 18.913025 | 3.303277e+07 | 0 | 0 | POINT (18.94092 -63.78413) | -0.019350 |
| 38244 | 6.714286 | 0.139510 | 1428857.875 | 0.026987 | 20 | 18.867090 | 18.901897 | 3.303278e+07 | 0 | 0 | POINT (18.9409 -63.78406) | -0.034807 |
| 38245 | 6.857143 | 0.135262 | 1397495.875 | 0.009361 | 20 | 18.852695 | 18.881191 | 3.303279e+07 | 0 | 0 | POINT (18.94089 -63.784) | -0.028496 |
38246 rows × 12 columns
Optical imagery to label point clouds#
Let’s use the Sentinel-2 satellite image we found to label each ATL07 point. We’ll
make a new column called sea_ice_label that can have either of these classifications:
thick/snow-covered sea ice
thin/gray sea ice
open water
These labels will be empirically determined based on the brightness (or surface reflectance) of the Sentinel-2 pixel.
Note
Sea ice can move very quickly in minutes, so while we’ve tried our best to find a Sentinel-2 image that was captured at about the same time as the ICESat-2 track, it is very likely that we will still be misclassifying some of the points below because the image and point clouds are not perfectly aligned.
# Get first STAC item from collection
item = item_collection.items[0]
item
- type "Feature"
- stac_version "1.0.0"
- id "S2B_2CND_20191031_0_L2A"
properties
- created "2022-11-08T11:22:41.199Z"
- platform "sentinel-2b"
- constellation "sentinel-2"
instruments[] 1 items
- 0 "msi"
- eo:cloud_cover 14.745366
- proj:epsg 32702
- mgrs:utm_zone 2
- mgrs:latitude_band "C"
- mgrs:grid_square "ND"
- grid:code "MGRS-2CND"
- view:sun_azimuth 48.0252837724571
- view:sun_elevation 25.759707849891
- s2:degraded_msi_data_percentage 0
- s2:nodata_pixel_percentage 36.36831
- s2:saturated_defective_pixel_percentage 0
- s2:dark_features_percentage 0
- s2:cloud_shadow_percentage 0
- s2:vegetation_percentage 0
- s2:not_vegetated_percentage 0.000125
- s2:water_percentage 85.254502
- s2:unclassified_percentage 0
- s2:medium_proba_clouds_percentage 4.393843
- s2:high_proba_clouds_percentage 4.110314
- s2:thin_cirrus_percentage 6.241209
- s2:snow_ice_percentage 0
- s2:product_type "S2MSI2A"
- s2:processing_baseline "02.13"
- s2:product_uri "S2B_MSIL2A_20191031T200529_N0213_R099_T02CND_20191031T221002.SAFE"
- s2:generation_time "2019-10-31T22:10:02.000000Z"
- s2:datatake_id "GS2B_20191031T200529_013853_N02.13"
- s2:datatake_type "INS-NOBS"
- s2:datastrip_id "S2B_OPER_MSI_L2A_DS_MTI__20191031T221002_S20191031T200532_N02.13"
- s2:granule_id "S2B_OPER_MSI_L2A_TL_MTI__20191031T221002_A013853_T02CND_N02.13"
- s2:reflectance_conversion_factor 1.01332239950472
- datetime "2019-10-31T20:05:54.244000Z"
- s2:sequence "0"
- earthsearch:s3_path "s3://sentinel-cogs/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A"
- earthsearch:payload_id "roda-sentinel2/workflow-sentinel2-to-stac/2a8eb3edebed97b8891482037ae45318"
- earthsearch:boa_offset_applied False
processing:software
- sentinel2-to-stac "0.1.0"
- updated "2022-11-08T11:22:41.199Z"
geometry
- type "Polygon"
coordinates[] 1 items
0[] 6 items
0[] 2 items
- 0 -171.0005820777167
- 1 -72.99570787454226
1[] 2 items
- 0 -167.92078532942014
- 1 -72.97252990541305
2[] 2 items
- 0 -168.93103098947427
- 1 -73.54545020350191
3[] 2 items
- 0 -169.73496041596468
- 1 -73.97622285629704
4[] 2 items
- 0 -171.00061679487823
- 1 -73.97992987556825
5[] 2 items
- 0 -171.0005820777167
- 1 -72.99570787454226
links[] 8 items
0
- rel "self"
- href "https://earth-search.aws.element84.com/v1/collections/sentinel-2-l2a/items/S2B_2CND_20191031_0_L2A"
- type "application/geo+json"
1
- rel "canonical"
- href "s3://sentinel-cogs/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/S2B_2CND_20191031_0_L2A.json"
- type "application/json"
2
- rel "license"
- href "https://sentinel.esa.int/documents/247904/690755/Sentinel_Data_Legal_Notice"
3
- rel "derived_from"
- href "https://earth-search.aws.element84.com/v1/collections/sentinel-2-l1c/items/S2B_2CND_20191031_0_L1C"
- type "application/geo+json"
4
- rel "parent"
- href "https://earth-search.aws.element84.com/v1/collections/sentinel-2-l2a"
- type "application/json"
5
- rel "collection"
- href "https://earth-search.aws.element84.com/v1/collections/sentinel-2-l2a"
- type "application/json"
6
- rel "root"
- href "https://earth-search.aws.element84.com/v1"
- type "application/json"
- title "Earth Search by Element 84"
7
- rel "thumbnail"
- href "https://earth-search.aws.element84.com/v1/collections/sentinel-2-l2a/items/S2B_2CND_20191031_0_L2A/thumbnail"
assets
aot
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/AOT.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Aerosol optical thickness (AOT)"
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
blue
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B02.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Blue (band 2) - 10m"
eo:bands[] 1 items
0
- name "blue"
- common_name "blue"
- description "Blue (band 2)"
- center_wavelength 0.49
- full_width_half_max 0.098
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
coastal
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B01.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Coastal aerosol (band 1) - 60m"
eo:bands[] 1 items
0
- name "coastal"
- common_name "coastal"
- description "Coastal aerosol (band 1)"
- center_wavelength 0.443
- full_width_half_max 0.027
- gsd 60
proj:shape[] 2 items
- 0 1830
- 1 1830
proj:transform[] 6 items
- 0 60
- 1 0
- 2 499980
- 3 0
- 4 -60
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 60
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
granule_metadata
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/granule_metadata.xml"
- type "application/xml"
roles[] 1 items
- 0 "metadata"
green
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B03.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Green (band 3) - 10m"
eo:bands[] 1 items
0
- name "green"
- common_name "green"
- description "Green (band 3)"
- center_wavelength 0.56
- full_width_half_max 0.045
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
nir
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B08.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "NIR 1 (band 8) - 10m"
eo:bands[] 1 items
0
- name "nir"
- common_name "nir"
- description "NIR 1 (band 8)"
- center_wavelength 0.842
- full_width_half_max 0.145
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
nir08
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B8A.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "NIR 2 (band 8A) - 20m"
eo:bands[] 1 items
0
- name "nir08"
- common_name "nir08"
- description "NIR 2 (band 8A)"
- center_wavelength 0.865
- full_width_half_max 0.033
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
nir09
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B09.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "NIR 3 (band 9) - 60m"
eo:bands[] 1 items
0
- name "nir09"
- common_name "nir09"
- description "NIR 3 (band 9)"
- center_wavelength 0.945
- full_width_half_max 0.026
- gsd 60
proj:shape[] 2 items
- 0 1830
- 1 1830
proj:transform[] 6 items
- 0 60
- 1 0
- 2 499980
- 3 0
- 4 -60
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 60
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
red
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B04.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Red (band 4) - 10m"
eo:bands[] 1 items
0
- name "red"
- common_name "red"
- description "Red (band 4)"
- center_wavelength 0.665
- full_width_half_max 0.038
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
rededge1
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B05.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Red edge 1 (band 5) - 20m"
eo:bands[] 1 items
0
- name "rededge1"
- common_name "rededge"
- description "Red edge 1 (band 5)"
- center_wavelength 0.704
- full_width_half_max 0.019
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
rededge2
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B06.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Red edge 2 (band 6) - 20m"
eo:bands[] 1 items
0
- name "rededge2"
- common_name "rededge"
- description "Red edge 2 (band 6)"
- center_wavelength 0.74
- full_width_half_max 0.018
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
rededge3
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B07.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Red edge 3 (band 7) - 20m"
eo:bands[] 1 items
0
- name "rededge3"
- common_name "rededge"
- description "Red edge 3 (band 7)"
- center_wavelength 0.783
- full_width_half_max 0.028
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
scl
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/SCL.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Scene classification map (SCL)"
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint8"
- spatial_resolution 20
roles[] 2 items
- 0 "data"
- 1 "reflectance"
swir16
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B11.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "SWIR 1 (band 11) - 20m"
eo:bands[] 1 items
0
- name "swir16"
- common_name "swir16"
- description "SWIR 1 (band 11)"
- center_wavelength 1.61
- full_width_half_max 0.143
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
swir22
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/B12.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "SWIR 2 (band 12) - 20m"
eo:bands[] 1 items
0
- name "swir22"
- common_name "swir22"
- description "SWIR 2 (band 12)"
- center_wavelength 2.19
- full_width_half_max 0.242
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
thumbnail
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/thumbnail.jpg"
- type "image/jpeg"
- title "Thumbnail image"
roles[] 1 items
- 0 "thumbnail"
tileinfo_metadata
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/tileinfo_metadata.json"
- type "application/json"
roles[] 1 items
- 0 "metadata"
visual
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/TCI.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "True color image"
eo:bands[] 3 items
0
- name "red"
- common_name "red"
- description "Red (band 4)"
- center_wavelength 0.665
- full_width_half_max 0.038
1
- name "green"
- common_name "green"
- description "Green (band 3)"
- center_wavelength 0.56
- full_width_half_max 0.045
2
- name "blue"
- common_name "blue"
- description "Blue (band 2)"
- center_wavelength 0.49
- full_width_half_max 0.098
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
roles[] 1 items
- 0 "visual"
wvp
- href "https://sentinel-cogs.s3.us-west-2.amazonaws.com/sentinel-s2-l2a-cogs/2/C/ND/2019/10/S2B_2CND_20191031_0_L2A/WVP.tif"
- type "image/tiff; application=geotiff; profile=cloud-optimized"
- title "Water vapour (WVP)"
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- unit "cm"
- scale 0.001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
aot-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/AOT.jp2"
- type "image/jp2"
- title "Aerosol optical thickness (AOT)"
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
blue-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B02.jp2"
- type "image/jp2"
- title "Blue (band 2) - 10m"
eo:bands[] 1 items
0
- name "blue"
- common_name "blue"
- description "Blue (band 2)"
- center_wavelength 0.49
- full_width_half_max 0.098
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
coastal-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B01.jp2"
- type "image/jp2"
- title "Coastal aerosol (band 1) - 60m"
eo:bands[] 1 items
0
- name "coastal"
- common_name "coastal"
- description "Coastal aerosol (band 1)"
- center_wavelength 0.443
- full_width_half_max 0.027
- gsd 60
proj:shape[] 2 items
- 0 1830
- 1 1830
proj:transform[] 6 items
- 0 60
- 1 0
- 2 499980
- 3 0
- 4 -60
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 60
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
green-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B03.jp2"
- type "image/jp2"
- title "Green (band 3) - 10m"
eo:bands[] 1 items
0
- name "green"
- common_name "green"
- description "Green (band 3)"
- center_wavelength 0.56
- full_width_half_max 0.045
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
nir-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B08.jp2"
- type "image/jp2"
- title "NIR 1 (band 8) - 10m"
eo:bands[] 1 items
0
- name "nir"
- common_name "nir"
- description "NIR 1 (band 8)"
- center_wavelength 0.842
- full_width_half_max 0.145
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
nir08-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B8A.jp2"
- type "image/jp2"
- title "NIR 2 (band 8A) - 20m"
eo:bands[] 1 items
0
- name "nir08"
- common_name "nir08"
- description "NIR 2 (band 8A)"
- center_wavelength 0.865
- full_width_half_max 0.033
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
nir09-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B09.jp2"
- type "image/jp2"
- title "NIR 3 (band 9) - 60m"
eo:bands[] 1 items
0
- name "nir09"
- common_name "nir09"
- description "NIR 3 (band 9)"
- center_wavelength 0.945
- full_width_half_max 0.026
- gsd 60
proj:shape[] 2 items
- 0 1830
- 1 1830
proj:transform[] 6 items
- 0 60
- 1 0
- 2 499980
- 3 0
- 4 -60
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 60
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
red-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B04.jp2"
- type "image/jp2"
- title "Red (band 4) - 10m"
eo:bands[] 1 items
0
- name "red"
- common_name "red"
- description "Red (band 4)"
- center_wavelength 0.665
- full_width_half_max 0.038
- gsd 10
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 10
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
rededge1-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B05.jp2"
- type "image/jp2"
- title "Red edge 1 (band 5) - 20m"
eo:bands[] 1 items
0
- name "rededge1"
- common_name "rededge"
- description "Red edge 1 (band 5)"
- center_wavelength 0.704
- full_width_half_max 0.019
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
rededge2-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B06.jp2"
- type "image/jp2"
- title "Red edge 2 (band 6) - 20m"
eo:bands[] 1 items
0
- name "rededge2"
- common_name "rededge"
- description "Red edge 2 (band 6)"
- center_wavelength 0.74
- full_width_half_max 0.018
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
rededge3-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B07.jp2"
- type "image/jp2"
- title "Red edge 3 (band 7) - 20m"
eo:bands[] 1 items
0
- name "rededge3"
- common_name "rededge"
- description "Red edge 3 (band 7)"
- center_wavelength 0.783
- full_width_half_max 0.028
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
scl-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/SCL.jp2"
- type "image/jp2"
- title "Scene classification map (SCL)"
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint8"
- spatial_resolution 20
roles[] 2 items
- 0 "data"
- 1 "reflectance"
swir16-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B11.jp2"
- type "image/jp2"
- title "SWIR 1 (band 11) - 20m"
eo:bands[] 1 items
0
- name "swir16"
- common_name "swir16"
- description "SWIR 1 (band 11)"
- center_wavelength 1.61
- full_width_half_max 0.143
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
swir22-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/B12.jp2"
- type "image/jp2"
- title "SWIR 2 (band 12) - 20m"
eo:bands[] 1 items
0
- name "swir22"
- common_name "swir22"
- description "SWIR 2 (band 12)"
- center_wavelength 2.19
- full_width_half_max 0.242
- gsd 20
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- scale 0.0001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
visual-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/TCI.jp2"
- type "image/jp2"
- title "True color image"
eo:bands[] 3 items
0
- name "red"
- common_name "red"
- description "Red (band 4)"
- center_wavelength 0.665
- full_width_half_max 0.038
1
- name "green"
- common_name "green"
- description "Green (band 3)"
- center_wavelength 0.56
- full_width_half_max 0.045
2
- name "blue"
- common_name "blue"
- description "Blue (band 2)"
- center_wavelength 0.49
- full_width_half_max 0.098
proj:shape[] 2 items
- 0 10980
- 1 10980
proj:transform[] 6 items
- 0 10
- 1 0
- 2 499980
- 3 0
- 4 -10
- 5 1900000
roles[] 1 items
- 0 "visual"
wvp-jp2
- href "s3://sentinel-s2-l2a/tiles/2/C/ND/2019/10/31/0/WVP.jp2"
- type "image/jp2"
- title "Water vapour (WVP)"
proj:shape[] 2 items
- 0 5490
- 1 5490
proj:transform[] 6 items
- 0 20
- 1 0
- 2 499980
- 3 0
- 4 -20
- 5 1900000
raster:bands[] 1 items
0
- nodata 0
- data_type "uint16"
- bits_per_sample 15
- spatial_resolution 20
- unit "cm"
- scale 0.001
- offset 0
roles[] 2 items
- 0 "data"
- 1 "reflectance"
bbox[] 4 items
- 0 -171.00061679487823
- 1 -73.97992987556825
- 2 -167.92078532942014
- 3 -72.97252990541305
stac_extensions[] 7 items
- 0 "https://stac-extensions.github.io/eo/v1.0.0/schema.json"
- 1 "https://stac-extensions.github.io/view/v1.0.0/schema.json"
- 2 "https://stac-extensions.github.io/mgrs/v1.0.0/schema.json"
- 3 "https://stac-extensions.github.io/raster/v1.1.0/schema.json"
- 4 "https://stac-extensions.github.io/processing/v1.1.0/schema.json"
- 5 "https://stac-extensions.github.io/grid/v1.0.0/schema.json"
- 6 "https://stac-extensions.github.io/projection/v1.0.0/schema.json"
- collection "sentinel-2-l2a"
Use stackstac.stack
to get the RGB bands from the Sentinel-2 image.
# Get RGB bands from Sentinel-2 STAC item as an xarray.DataArray
da_image = stackstac.stack(
items=item,
assets=["red", "green", "blue"],
resolution=60,
rescale=False, # keep original 14-bit scale
dtype=np.float32,
fill_value=np.float32(np.nan),
)
da_image = da_image.squeeze(dim="time") # drop time dimension so only (band, y, x) left
da_image
<xarray.DataArray 'stackstac-bd9892c19b6c86ec2d80ee484fd3c349' (band: 3,
y: 1831, x: 1830)> Size: 40MB
dask.array<getitem, shape=(3, 1831, 1830), dtype=float32, chunksize=(1, 1024, 1024), chunktype=numpy.ndarray>
Coordinates: (12/54)
time datetime64[ns] 8B 2019-10-31T20:...
id <U23 92B 'S2B_2CND_20191031_0_L2A'
* band (band) <U5 60B 'red' 'green' 'blue'
* x (x) float64 15kB 5e+05 ... 6.097...
* y (y) float64 15kB 1.9e+06 ... 1.7...
s2:sequence <U1 4B '0'
... ...
title (band) <U20 240B 'Red (band 4) -...
raster:bands object 8B {'nodata': 0, 'data_ty...
common_name (band) <U5 60B 'red' 'green' 'blue'
center_wavelength (band) float64 24B 0.665 0.56 0.49
full_width_half_max (band) float64 24B 0.038 ... 0.098
epsg int64 8B 32702
Attributes:
spec: RasterSpec(epsg=32702, bounds=(499980, 1790160, 609780, 1900...
crs: epsg:32702
transform: | 60.00, 0.00, 499980.00|\n| 0.00,-60.00, 1900020.00|\n| 0.0...
resolution: 60# Get bounding box of Sentinel-2 image to spatially subset ATL07 geodataframe
bbox = shapely.geometry.box(*da_image.rio.bounds()) # xmin, ymin, xmax, ymax
gdf = gdf.to_crs(crs=da_image.crs) # reproject point cloud to Sentinel-2 image's CRS
gdf = gdf[gdf.within(other=bbox)].reset_index() # subset point cloud to Sentinel-2 bbox
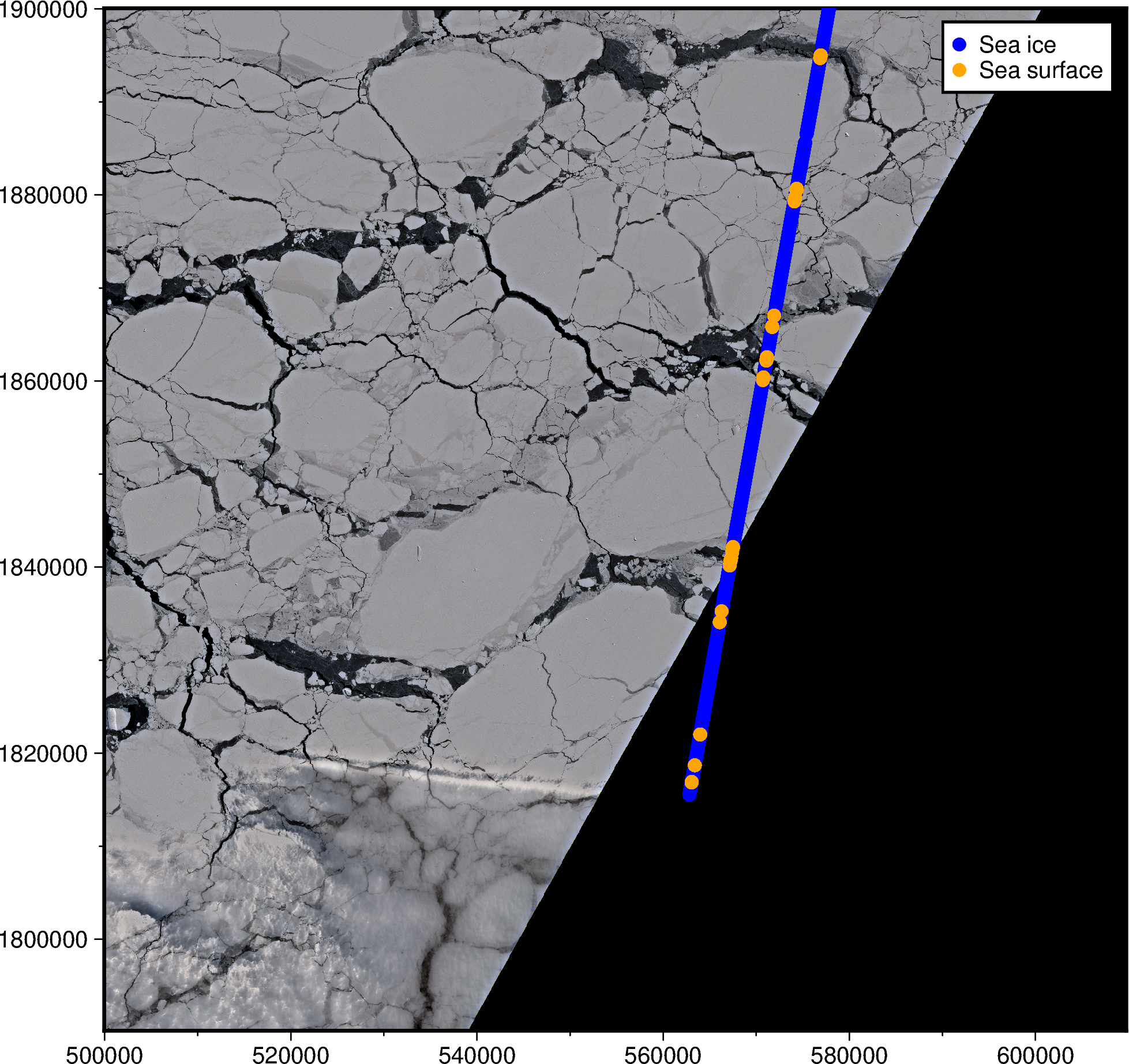
Let’s plot the Sentinel-2 satellite image and ATL07 points! The ATL07 data has a
height_segment_ssh_flag variable with a binary sea ice (0) / sea surface (1)
classification, which we’ll plot using two different colors to check that things
overlap more or less.
fig = pygmt.Figure()
# Plot Sentinel-2 RGB image
# Convert from 14-bit to 8-bit color scale for PyGMT
fig.grdimage(grid=((da_image / 2**14) * 2**8).astype(np.uint8), frame=True)
# Plot ATL07 points
# Sea ice points in blue
df_ice = gdf[gdf.height_segment_ssh_flag == 0].get_coordinates()
fig.plot(x=df_ice.x, y=df_ice.y, style="c0.2c", fill="blue", label="Sea ice")
# Sea surface points in orange
df_water = gdf[gdf.height_segment_ssh_flag == 1].get_coordinates()
fig.plot(x=df_water.x, y=df_water.y, style="c0.2c", fill="orange", label="Sea surface")
fig.legend()
fig.show()
Looking good! Notice how the orange (sea surface) coincide with the cracks in the sea ice.
Next, we’ll want to do better than a binary 0/1 or ice/water classification, and pick up different shades of gray (white thick ice, gray thin ice, black water). Let’s use the X and Y coordinates from the point cloud to pick up surface reflectance values from the Sentinel-2 image.
PyGMT’s grdtrack
function can be used to do this X/Y sampling from a 1-band grid.
df_red = pygmt.grdtrack(
grid=da_image.sel(band="red").compute(), # Choose only the Red band
points=gdf.get_coordinates(), # x/y coordinates from ATL07
newcolname="red_band_value",
interpolation="n", # nearest neighbour
)
grdtrack [WARNING]: Some input points were outside the grid domain(s).
# Plot cross-section
df_red.plot.scatter(
x="y", y="red_band_value", title="Sentinel-2 red band values in y-direction"
)
<Axes: title={'center': 'Sentinel-2 red band values in y-direction'}, xlabel='y', ylabel='red_band_value'>
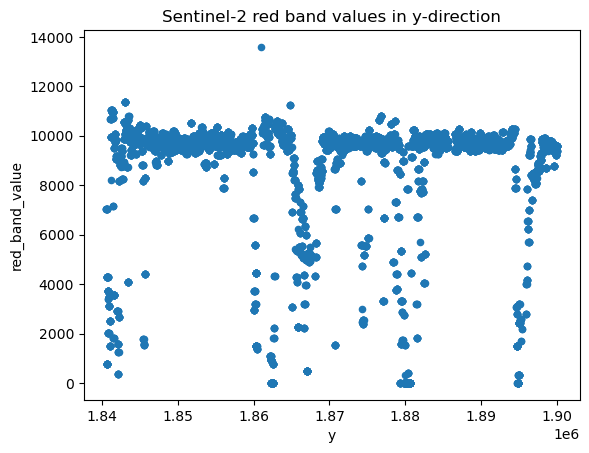
The cross-section view shows most points having a Red band reflectance value of 10000, that should correspond to white sea ice. Darker values near 0 would be water, and intermediate values around 6000 would be thin ice.
(Click ‘Show code cell content’ below if you’d like to see the histogram plot)
Show code cell content
df_red.plot(
kind="hist", column="red_band_value", bins=30, title="Sentinel-2 red band values"
)
<Axes: title={'center': 'Sentinel-2 red band values'}, ylabel='Frequency'>
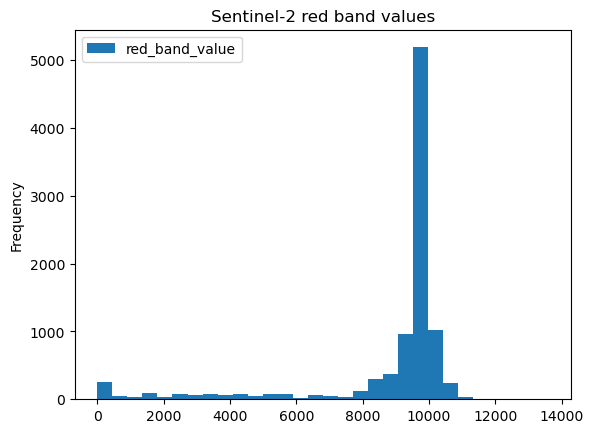
To keep things simple, we’ll label the surface_type of each ATL07 point
using a simple threshold:
Int label |
Surface Type |
Bin values |
|---|---|---|
0 |
Water (dark) |
|
1 |
Thin sea ice (gray) |
|
2 |
Thick sea ice (white) |
|
gdf["surface_type"] = pd.cut(
x=df_red["red_band_value"],
bins=[0, 4000, 8000, 14000],
labels=[0, 1, 2], # "water", "thin_ice", "thick_ice"
)
There are some NaN values in some rows of our geodataframe (which had no matching Sentinel-2 pixel value) that should be dropped here.
gdf = gdf.dropna().reset_index(drop=True)
gdf
| index | photon_rate | hist_w | background_r_norm | height_segment_height | height_segment_n_pulse_seg | hist_mean_h | hist_median_h | x_atc | layer_flag | height_segment_ssh_flag | geometry | hist_mean_median_h_diff | surface_type | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 4388 | 8.647058 | 0.224312 | 5565264.00 | 0.605202 | 16 | -67.327896 | -67.305969 | 2.821440e+07 | 0 | 0 | POINT (577809.769 1900049.731) | -0.021927 | 2 |
| 1 | 4389 | 8.058824 | 0.173039 | 5587664.50 | 0.669868 | 16 | -67.209755 | -67.219963 | 2.821441e+07 | 0 | 0 | POINT (577808.777 1900044.162) | 0.010208 | 2 |
| 2 | 4390 | 7.941176 | 0.190458 | 5587664.50 | 0.736938 | 16 | -67.143631 | -67.138664 | 2.821441e+07 | 0 | 0 | POINT (577807.757 1900038.423) | -0.004967 | 2 |
| 3 | 4391 | 8.294118 | 0.250401 | 5587664.50 | 0.701096 | 16 | -67.213936 | -67.217377 | 2.821442e+07 | 0 | 0 | POINT (577806.783 1900032.932) | 0.003441 | 2 |
| 4 | 4392 | 7.882353 | 0.170319 | 5587670.50 | 0.620872 | 16 | -67.279953 | -67.277100 | 2.821442e+07 | 0 | 0 | POINT (577805.803 1900027.401) | -0.002853 | 2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 9449 | 13837 | 8.750000 | 0.148517 | 3209719.50 | -0.032860 | 15 | -68.235619 | -68.211128 | 2.827476e+07 | 0 | 0 | POINT (567235.505 1840650.409) | -0.024490 | 1 |
| 9450 | 13838 | 9.866667 | 0.113973 | 3322385.75 | -0.038391 | 14 | -68.211784 | -68.208221 | 2.827476e+07 | 0 | 0 | POINT (567234.707 1840645.805) | -0.003563 | 1 |
| 9451 | 13839 | 11.461538 | 0.130621 | 3322385.75 | -0.046043 | 12 | -68.220390 | -68.207855 | 2.827477e+07 | 0 | 1 | POINT (567233.997 1840641.703) | -0.012535 | 1 |
| 9452 | 13840 | 12.083333 | 0.133239 | 3322386.75 | -0.070625 | 11 | -68.228592 | -68.216240 | 2.827477e+07 | 0 | 1 | POINT (567233.382 1840638.147) | -0.012352 | 1 |
| 9453 | 13841 | 10.500000 | 0.136830 | 3322399.50 | -0.038437 | 13 | -68.216888 | -68.200233 | 2.827478e+07 | 0 | 0 | POINT (567232.672 1840634.043) | -0.016655 | 1 |
9454 rows × 14 columns
Save to GeoParquet#
Let’s save the ATL07 point cloud data to a GeoParquet file so we don’t have to run all the pre-processing code above again.
gdf.to_parquet(path="ATL07_point_cloud.gpq", compression="zstd", schema_version="1.1.0")
Note
To compress or not? When storing your data, note that there is a tradeoff in terms of compression and read speeds. Uncompressed data would typically be fastest to read (assuming no network transfer) but result in large file sizes. We’ll choose Zstandard (zstd) as the compression method here as it provides a balance between fast reads (quicker than the default ‘snappy’ compression codec), and good compression into a small file size.
# Load GeoParquet file back into geopandas.GeoDataFrame
gdf = gpd.read_parquet(path="ATL07_point_cloud.gpq")
Part 2: DataLoader and Model architecture#
The following parts will bring us one step closer to having a full machine learning pipeline. We will create:
A ‘DataLoader’, which is a fancy data container we can loop over; and
A neural network ‘model’ that will take our input ATL07 data and output point cloud classifications.
From dataframe tables to batched tensors#
Machine learning models are compute intensive, and typically run on specialized hardware called Graphical Processing Units (GPUs) instead of ordinary CPUs. Depending on your input data format (images, tables, audio, etc), and the machine learning library/framework you’ll use (e.g. Pytorch, Tensorflow, RAPIDS AI CuML, etc), there will be different ways to transfer data from disk storage -> CPU -> GPU.
For this exercise, we’ll be using PyTorch, and do the following data conversions:
geopandas.GeoDataFrame ->
pandas.DataFrame ->
torch.Tensor ->
torch Dataset ->
torch DataLoader
# Select data variables from DataFrame that will be used for training
df = gdf[
[
# Input variables
"photon_rate",
"hist_w",
"background_r_norm",
"height_segment_height",
"height_segment_n_pulse_seg",
"hist_mean_median_h_diff",
# Output label (groundtruth)
"surface_type",
]
]
tensor = torch.from_numpy( # convert pandas.DataFrame to torch.Tensor (via numpy)
df.to_numpy(dtype="float32")
)
# assert tensor.shape == torch.Size([9454, 7]) # (rows, columns)
dataset = torch.utils.data.TensorDataset(tensor) # turn torch.Tensor into torch Dataset
dataloader = torch.utils.data.DataLoader( # put torch Dataset in a DataLoader
dataset=dataset,
batch_size=128, # mini-batch size
shuffle=True,
)
This PyTorch
DataLoader
can be used in a for-loop to produce mini-batch tensors of shape (128, 7) later below.
Choosing a Machine Learning algorithm#
Next is to pick a supervised learning ‘model’ for our point cloud classification task. There are a variety of machine learning methods to choose with different levels of complexity:
Easy - Decision trees (e.g. XGBoost, Random Forest), K-Nearest Neighbors, etc
Medium - Basic neural networks (e.g. Multi-layer Perceptron, Convolutional neural networks, etc).
Hard - State-of-the-art models (e.g. Graph Neural Networks, Transformers, State Space Models)
Let’s take the middle ground and build a multi-layer perceptron, also known as an artificial feedforward neural network.
See also
There are many frameworks catering to the different levels of Machine Learning models mentioned above. Some notable ones are:
Easy: ‘Classic’ ML - Scikit-learn (CPU-based) and CuML (GPU-based)
Medium: DIY Neural networks - Pytorch and Tensorflow
Hard: High-level ML frameworks - Lightning, HuggingFace, etc.
While you might think that going from easy to hard is recommended, there are some people who actually start with a (well-documented) framework and work their way down! Do whatever works best for you on your machine learning journey.
A Pytorch model or
torch.nn.Module is
constructed as a Python class with an __init__ method for the neural network layers,
and a forward method for the forward pass (how the data passes through the layers).
This multi-layer perceptron below will have:
An input layer with 6 nodes, corresponding to the 6 input data variables
Two hidden layers, 50 nodes each
Output layer with 3 nodes, for 3 surface types (open water, thin ice, thick/snow-covered ice)
class PointCloudClassificationModel(torch.nn.Module):
def __init__(self):
super().__init__()
self.linear1 = torch.nn.Linear(in_features=6, out_features=50)
self.linear2 = torch.nn.Linear(in_features=50, out_features=50)
self.linear3 = torch.nn.Linear(in_features=50, out_features=3)
def forward(self, x: torch.Tensor) -> torch.Tensor:
x1 = self.linear1(x)
x2 = self.linear2(x1)
x3 = self.linear3(x2)
return x3
model = PointCloudClassificationModel()
# model = model.to(device="cuda") # uncomment this line if running on GPU
model
PointCloudClassificationModel(
(linear1): Linear(in_features=6, out_features=50, bias=True)
(linear2): Linear(in_features=50, out_features=50, bias=True)
(linear3): Linear(in_features=50, out_features=3, bias=True)
)
Part 3: Training the neural network#
Now is the time to train the ML model! We’ll need to:
Choose a loss function and optimizer
Configure training hyperparameters such as the learning rate (
lr) and number of epochs (max_epochs) or iterations over the entire training dataset.Construct the main training loop to:
get a mini-batch from the DataLoader
pass the mini-batch data into the model to get a prediction
minimize the error (or loss) between the prediction and groundtruth
Let’s see how this is done!
# Setup loss function and optimizer
loss_bce = torch.nn.BCEWithLogitsLoss() # binary cross entropy loss
optimizer = torch.optim.Adam(params=model.parameters(), lr=0.001)
# Main training loop
max_epochs: int = 3
size = len(dataloader.dataset)
for epoch in tqdm.tqdm(iterable=range(max_epochs)):
for i, batch in enumerate(dataloader):
minibatch: torch.Tensor = batch[0]
# assert minibatch.shape == (128, 7)
assert minibatch.device == torch.device("cpu") # Data is on CPU
# Uncomment two lines below if GPU is available
# minibatch = minibatch.to(device="cuda") # Move data to GPU
# assert minibatch.device == torch.device("cuda:0") # Data is on GPU now!
# Split data into input (x) and target (y)
x = minibatch[:, :6] # Input is in first 6 columns
y = minibatch[:, 6] # Output (groundtruth) is in 7th column
y_target = torch.nn.functional.one_hot(y.to(dtype=torch.int64), 3) # 3 classes
# Pass data into neural network model
prediction = model(x=x)
# Compute prediction error
loss = loss_bce(input=prediction, target=y_target.to(dtype=torch.float32))
# Backpropagation (to minimize loss)
loss.backward()
optimizer.step()
optimizer.zero_grad()
# Report metrics
current = (i + 1) * len(x)
print(f"loss: {loss:>7f} [{current:>5d}/{size:>5d}]")
67%|██████▋ | 2/3 [00:00<00:00, 8.63it/s]
loss: 7949.509277 [ 8140/ 9454]
loss: 3678.343994 [ 8140/ 9454]
100%|██████████| 3/3 [00:00<00:00, 8.64it/s]
loss: 1735.765503 [ 8140/ 9454]
Did the model learn something? A good sign to check is if the loss value is decreasing, which means the error between the predicted and groundtruth value is getting smaller.
Inference results#
Besides monitoring the loss value, it is also good to calculate a metric like Precision, Recall or F1-score. Let’s first run the model in ‘inference’ mode to get predictions.
gdf["predicted_surface_type"] = None # create new column with NaN to store results
with torch.inference_mode():
for i, batch in enumerate(dataloader):
minibatch: torch.Tensor = batch[0]
x = minibatch[:, :6]
prediction = model(x=x) # one-hot encoded predictions
prediction_labels = torch.argmax(input=prediction, dim=1) # 0/1/2 labels
start_index = i * dataloader.batch_size
stop_index = start_index + len(minibatch) - 1
gdf.loc[start_index:stop_index, "predicted_surface_type"] = prediction_labels
Caution
Ideally, you would want to run inference on a hold-out validation or test set, rather
than the points the model was trained on! See e.g.
sklearn.model_selection.train_test_split
on how this can be done.
Now that we have the predicted results in the predicted_surface_type column, we can
compare it with the ‘groundtruth’ labels in the ‘surface_type’ column by visualizing
it in a confusion matrix.
pd.crosstab(
index=gdf.surface_type,
columns=gdf.predicted_surface_type,
rownames=["Actual"],
colnames=["Predicted"],
margins=True,
)
| Predicted | 1 | 2 | All |
|---|---|---|---|
| Actual | |||
| 0 | 0 | 725 | 725 |
| 1 | 1 | 577 | 578 |
| 2 | 2 | 8149 | 8151 |
| All | 3 | 9451 | 9454 |
Attention
Oo, it looks like our model isn’t producing very good results! It’s practically only predicting thick sea ice (class: 2). There could be many reasons for this, and it’s up to you to figure out a solution, either by changing the data, or adjusting the model.
Data-centric approaches:
Add more data! Maybe <10000 points isn’t enough, try getting more!
Check the labels! Are there wrongly labelled points? Is the Sentinel-2 dark/gray/bright bins above too simplistic? Investigate!
Normalize the data value range. The original paper by Koo et al., 2023 applied min-max normalization on the 6 input columns, try and apply that too!
Model-centric appraoches:
Manage class imbalance. There are a lot more thick sea ice points than thin sea ice or water points, could we modify the loss function to weigh rare classes higher?
Adjust the model hyperparameters, try adjusting the learning rate, train the model for more epochs, etc.
Tweak the model architecture. The original paper by Koo et al., 2023 used a
tanhactivation function in the neural network layers. Will adding that help?
The list above isn’t exhaustive, and different machine learning practicioners may have other suggestions on what to try next. That said, you now have a Machine Learning ready GeoParquet dataset to iterate on ideas more quickly. Good luck!
References#
Koo, Y., Xie, H., Kurtz, N. T., Ackley, S. F., & Wang, W. (2023). Sea ice surface type classification of ICESat-2 ATL07 data by using data-driven machine learning model: Ross Sea, Antarctic as an example. Remote Sensing of Environment, 296, 113726. https://doi.org/10.1016/j.rse.2023.113726
Petty, A. A., Bagnardi, M., Kurtz, N. T., Tilling, R., Fons, S., Armitage, T., Horvat, C., & Kwok, R. (2021). Assessment of ICESat‐2 Sea Ice Surface Classification with Sentinel‐2 Imagery: Implications for Freeboard and New Estimates of Lead and Floe Geometry. Earth and Space Science, 8(3), e2020EA001491. https://doi.org/10.1029/2020EA001491